Weissleder Lab
Read BioThe lab's overall goal is to:
- Obtain a deeper understanding of human biology in health and disease
- Translate new biological understanding into clinically useful diagnostics
- Identify new therapeutic approaches and drug targets
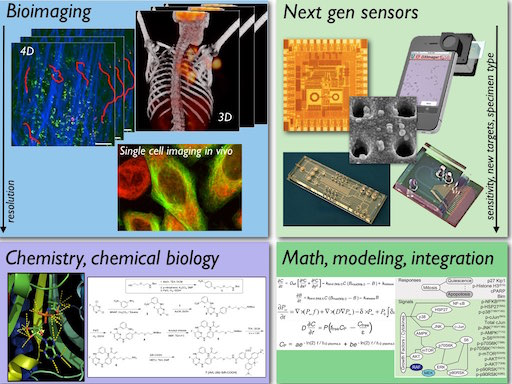
For some of the current research areas please see here. We develop and use a variety of cutting-edge approaches to discover new biology. Some of these include advanced imaging, novel devices and sensor technologies, chemical biology and modeling. In general, we aim at obtaining:
- Quantitative in vivo measurements
- Systems-wide global measurements
- Dynamic, serial measurements
- A fine-grained understanding of in vivo function
We strongly encourage interdisciplinary approaches, fostering collaboration between scientists and clinicians from diverse backgrounds. This convergence approach has led to many exciting discoveries and innovations at the Weissleder Lab.
Recent Publications
Nahrendorf M
Recognizing Early Career Translational Investigators. JACC Basic Transl Sci. 2026;11(1):101467 - PMID: 41589777 - PMCID: PMC12902213 - DOI: 10.1016/j.jacbts.2025.101467O'Shea A, Lewis AJM, Caravan P, Izquierdo-Garcia D, Le Fur M, Catalano OA, Montesi SB, Quintana JM, Carlson JCT, Dubach JM, Castro CM, Bobić M, Huesa-Berral C, Ng TSC, Bertolet A, Nahrendorf M, Weissleder R
First-in-Human Study of the Carbohydrate Nanoparticle 64Cu-Macrin. J Nucl Med. 2025;:ePub - PMID: 41469153 - DOI: 10.2967/jnumed.125.270770Zitti B, Duval F, Wirapati P, Hicham M, Xie Y, Oh J, Hoelzl J, Meiser P, Varrone M, Peterson HM, Cianciaruso C, Bill R, Bayerl F, Bolli E, Goubet AG, Kiss M, McDowell S, Cheng P, Celestini D, Terzic J, Zwahlen T, Alouche N, Zouggari N, Tarussio D, Tissot S, Nunes-Hasler P, Mino-Kenudson M, Lanuti M, Faquin WC, Sadow PM, Tille JC, Intidhar Labidi-Galy S, Garris CS, Hugues S, Petrova TV, Ludewig B, Quezada S, Luther S, Mempel TR, Ciriello G, Pai SI, Michielin O, Böttcher JP, Weissleder R, Pittet MJ
Positioning and reversible suppression of CCR7+ dendritic cells in perivascular tumor niches shape cancer immunity. Immunity. 2025;59(1):161-176.e12 - PMID: 41421339 - PMCID: PMC12882814 - DOI: 10.1016/j.immuni.2025.11.020Garris C, Dunn GP, Weissleder R
Targeting myeloid cell polarization to prevent GBM recurrence. Mol Ther. 2025;34(1):41-43 - PMID: 41421347 - DOI: 10.1016/j.ymthe.2025.12.019Choi M, Jeong MH, Son T, Kwak TJ, Jo JH, Nahm JH, Weissleder R, Jang SI, Im H
Integrated Multi-Channel Automatic Cellular Profiling (iMAP) System for Cholangiocarcinoma Diagnosis. Small. 2025;22(2):e07120 - PMID: 41392743 - PMCID: PMC12747514 - DOI: 10.1002/smll.202507120- More publications ...
Research projects
We select research ideas and topics that have the greatest positive impact in medicine and biology. We attract intensely inquisitive and motivated trainees and embrace inter- and neodisciplinary approaches to sustain a high degree of innovation. Some of the past accomplishments can be found here. Some current research projects include:
Single and scant cell analysis
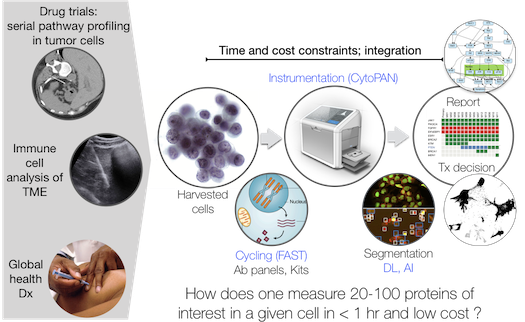 Cellular cancer diagnostics are essential to establish the correct diagnosis, choose appropriate treatments, enroll patients in experimental trials, assess therapeutic efficacy or re-stage disease—in short, all clinical decision making steps. Often obtained by image guided intervention, current workflows of tissue processing remain labor intensive, time consuming, limited in diagnostic read-outs, morbidity prone, costly and are not infallible. Automated, rapid on site assessment of cellular rather than tissue specimens by fine needle aspiration (FNA) could circumvent these bottlenecks. We have developed several new multiplex cycling techniques for single and scant cell analysis including ABCD (Science Transl Medicine. 2014;6:219ra9), SCANT (Nat Commun. 2018;9:4550) and FAST. The FAST method is based on new chemistry of site specific clickable dyes and quenchers (Carlson et al., 2018, J Am Chem Soc, 140, 3603-3612; Carlson et al., 2013, Angew Chem Int Ed Engl, 52, 6917-20), is ultrafast and highly mutliplexable. Compared to existing technology (immunohistochemistry, flow cytometry and mass spectrometry) it: i) allows dozens of markers to be analyzed in an hour, ii) works well in single cells or small numbers of cells, iii) is fast and inexpensive and iv) can be combined with genomic analytical techniques. These technologies, together with scRNAseq, are currently used to investigate questions such as: i) How divergent are protein profiles within a malignant lesion (intra-tumor heterogeneity) and between patients (inter-patient heterogeneity)? ii) are drug targets and pathways being modulated with targeted therapies? iii) What does the immune cell make-up change temporally duding immunotherapy in patients? iv) are there currently undefined innate immune cells populations in human patient samples that could be targeted therapeutically? and v) can the technologies be used for global health applications in low and middle income countries (LMIC)?
Cellular cancer diagnostics are essential to establish the correct diagnosis, choose appropriate treatments, enroll patients in experimental trials, assess therapeutic efficacy or re-stage disease—in short, all clinical decision making steps. Often obtained by image guided intervention, current workflows of tissue processing remain labor intensive, time consuming, limited in diagnostic read-outs, morbidity prone, costly and are not infallible. Automated, rapid on site assessment of cellular rather than tissue specimens by fine needle aspiration (FNA) could circumvent these bottlenecks. We have developed several new multiplex cycling techniques for single and scant cell analysis including ABCD (Science Transl Medicine. 2014;6:219ra9), SCANT (Nat Commun. 2018;9:4550) and FAST. The FAST method is based on new chemistry of site specific clickable dyes and quenchers (Carlson et al., 2018, J Am Chem Soc, 140, 3603-3612; Carlson et al., 2013, Angew Chem Int Ed Engl, 52, 6917-20), is ultrafast and highly mutliplexable. Compared to existing technology (immunohistochemistry, flow cytometry and mass spectrometry) it: i) allows dozens of markers to be analyzed in an hour, ii) works well in single cells or small numbers of cells, iii) is fast and inexpensive and iv) can be combined with genomic analytical techniques. These technologies, together with scRNAseq, are currently used to investigate questions such as: i) How divergent are protein profiles within a malignant lesion (intra-tumor heterogeneity) and between patients (inter-patient heterogeneity)? ii) are drug targets and pathways being modulated with targeted therapies? iii) What does the immune cell make-up change temporally duding immunotherapy in patients? iv) are there currently undefined innate immune cells populations in human patient samples that could be targeted therapeutically? and v) can the technologies be used for global health applications in low and middle income countries (LMIC)?
- Ko J, Oh J, Ahmed MS, Carlson JCT, Weissleder R. Ultra-fast cycling for multiplexed cellular fluorescence imaging. Angew Chem IE. 2019; in review.
- Giedt RJ, Pathania D, Carlson JCT, McFarland PJ, Del Castillo AF, Juric D, Weissleder R. Single-cell barcode analysis provides a rapid readout of cellular signaling pathways in clinical specimens. Nat Commun. 2018;9(1):4550 - PMID: 30382095
- Im H, Pathania D, McFarland PJ, Sohani AR, Degani I, Allen M, Coble M, Kilcoyne A, Hong S, Rohrer L, Abramson JS, Dryden-Peterson S, Fexon L, Pivovarov M, Chabner B, Lee H, Castro CM, Weissleder R. Design and clinical validation of a point-of-care device for the diagnosis of lymphoma via contrast- enhanced microholography and machine learning. Nat Biomed Eng. 2018;2(9):666-674 - PMID: 30555750
- Weissleder R, Lee H. Automated molecular image cytometry and analysis in modern oncology. Nat Rev Mat 2020, in press.
Single Exosome analysis
 Exosomes (and other extracelluar vesicles, EVs) are shed into circulation by most cancer cells and afford an opportunity to longitudinally study tumor evolution and (non)response to therapies in realtime. Despite the relative abundance of tumor-derived EVs and their informative cargo, one of the major bottlenecks to their use as biomarkers has been diagnostic sensitivity. We have developed a number of different exosome analyses platforms and are focusing on single exosome analysis. In addition to serving as a biomarker discovery platform, analysis of different EVs will provide insight into how many different types of tumor EVs there are, whether EV produced by host cells in tumors can be detected in circulation and how EV change the phenotype during treatment.
Exosomes (and other extracelluar vesicles, EVs) are shed into circulation by most cancer cells and afford an opportunity to longitudinally study tumor evolution and (non)response to therapies in realtime. Despite the relative abundance of tumor-derived EVs and their informative cargo, one of the major bottlenecks to their use as biomarkers has been diagnostic sensitivity. We have developed a number of different exosome analyses platforms and are focusing on single exosome analysis. In addition to serving as a biomarker discovery platform, analysis of different EVs will provide insight into how many different types of tumor EVs there are, whether EV produced by host cells in tumors can be detected in circulation and how EV change the phenotype during treatment.
- Yang KS*, Im H*, Hong S, Pergolini I, Del Castillo AF, Wang R, Clardy S, Huang CH, Pille C, Ferrone S, Yang R, Castro CM, Lee H, Del Castillo CF, Weissleder R. Multiparametric plasma EV profiling facilitates diagnosis of pancreatic malignancy. Sci Transl Med. 2017;9(391):eaal3226.
- Ko J, Wang Y, Gungabeesoon J, Pittet M, Weitz D, Weissleder R. Droplet-based single EV sequencing for rare immune subtype discovery. µ-TAS 2019. 2019; T104c 85.
- Shao H, Im H, Castro CM, Breakefield X, Weissleder R, Lee H. New Technologies for Analysis of Extracellular Vesicles. Chem Rev. 2018; 118(4) 1917-1950.
- Fraser K, Jo A, Giedt J et al. Characterization of single microvesicles in plasma from glioblastoma patients. Neuro Oncol. 2019; 21(5) 606-615. doi:10.1093/neuonc/noy187.
Immunotherapeutics and innate immune cell function
 Tumor-associated macrophages (TAM) are often abundant in the tumor microenvironment and play important roles during tumor spread and in response to therapy. TAM can show considerable plasticity by assuming phenotypes and functions that are either tumoricidal (e.g. M1-like cells) or tumorigenic (e.g. M2-like cells). TAM can also profoundly influence the efficacy of anticancer drugs, nanotherapeutics and immunotherapeutics. Furthermore, TAM have an increasingly well-documented role as relevant therapeutic targets, for example through targeting of the colony-stimulating factor 1 receptor (CSF1R). However, most of our knowledge on TAM and other tumor-associated myeloid cells comes from histological examinations, ex vivo flow cytometry, transcriptome profiling or in vitro culture experiments. There is a significant knowledge gap on how these cells function in vivo. One current project uses high resolution imaging to map TAM subset activity during tumor progression and defines how TAM-targeting agents alter this activity. Related projects are aimed at understanding efficacy or resistance to anti-PD1, anti-PDL1 and anti-CTLA4 therapies.
Tumor-associated macrophages (TAM) are often abundant in the tumor microenvironment and play important roles during tumor spread and in response to therapy. TAM can show considerable plasticity by assuming phenotypes and functions that are either tumoricidal (e.g. M1-like cells) or tumorigenic (e.g. M2-like cells). TAM can also profoundly influence the efficacy of anticancer drugs, nanotherapeutics and immunotherapeutics. Furthermore, TAM have an increasingly well-documented role as relevant therapeutic targets, for example through targeting of the colony-stimulating factor 1 receptor (CSF1R). However, most of our knowledge on TAM and other tumor-associated myeloid cells comes from histological examinations, ex vivo flow cytometry, transcriptome profiling or in vitro culture experiments. There is a significant knowledge gap on how these cells function in vivo. One current project uses high resolution imaging to map TAM subset activity during tumor progression and defines how TAM-targeting agents alter this activity. Related projects are aimed at understanding efficacy or resistance to anti-PD1, anti-PDL1 and anti-CTLA4 therapies.
- Weissleder R, Pittet M. A new worldview of inflammatory cells affecting cancer therapy. Nat Biomed Eng. 2020; in press.
- Rodell CB, Arlauckas SP, Cuccarese MF et al. TLR7/8-agonist-loaded nanoparticles promote the polarization of tumour-associated macrophages to enhance cancer immunotherapy. Nat Biomed Eng. 2018; 2(8) 578-588.
- Arlauckas SP*, Garris CS*, Kohler RH, Kitaoka M, Cuccarese MF, Yang KS, Miller MA, Carlson JC, Freeman GJ, Anthony RM, Weissleder R, Pittet MJ, In vivo imaging reveals a tumor-associated macrophage–mediated resistance pathway in anti–PD-1 therapy. Sci Transl Med. 2017;9(389):eaal3604 – PMID: 28490665 – PMCID: PMC5734617 – DOI: 10.1126/scitranslmed.aal3604.
- Engblom C*, Pfirschke C*, Zilionis R, Da Silva Martins J, Bos SA, Courties G, Rickelt S, Severe N, Baryawno N, Faget J, Savova V, Zemmour D, Kline J, Siwicki M, Garris C, Pucci F, Liao HW, Lin YJ, Newton A, Yaghi OK, Iwamoto Y, Tricot B, Wojtkiewicz GR, Nahrendorf M, Cortez-Retamozo V, Meylan E, Hynes RO, Demay M, Klein A, Bredella MA, Scadden DT, Weissleder R, Pittet MJ. Osteoblasts remotely supply lung tumors with cancer-promoting SiglecFhigh neutrophils. Science. 2017;358:eaal5081 – PMID: 29191879 – DOI: 10.1126/science.aal5081
- Rodell CB, Koch PD, Weissleder R. Screening for new macrophage therapeutics. Theranostics. 2019; 9(25) 7714-7729.
Systems pharmacology
 We use the term “systems pharmacology” to define research that aims at understanding how drugs work (on specific pathways, on different cell types and in different tissues/organs/diseases), what the variability in patient response is and why many cancer treatments fail. Using a broad array of technologies including in vivo imaging of pharmacokinetics, intravital pharmacodynamic imaging (IPDI), mass spectrometry analysis, novel nanotechnology sensing approaches and chemical biology we obtain quantitative measurements and then develop mechanistic and probabilistic models. Network analysis and quantitative measurements of drug actions and side effects play a key component. Ultimately, we hope to improve our poor understanding of treatment response and define new targets drugs (or synergistic combinations) to tackle complex diseases.
We use the term “systems pharmacology” to define research that aims at understanding how drugs work (on specific pathways, on different cell types and in different tissues/organs/diseases), what the variability in patient response is and why many cancer treatments fail. Using a broad array of technologies including in vivo imaging of pharmacokinetics, intravital pharmacodynamic imaging (IPDI), mass spectrometry analysis, novel nanotechnology sensing approaches and chemical biology we obtain quantitative measurements and then develop mechanistic and probabilistic models. Network analysis and quantitative measurements of drug actions and side effects play a key component. Ultimately, we hope to improve our poor understanding of treatment response and define new targets drugs (or synergistic combinations) to tackle complex diseases.
Some of the Questions we are currently addressing are:
- What are the molecular and cellular pathways from drug engaging target to tumor regression for any drug ?
- What makes a cancer cell more, or less, responsive to drug X than a cell in the most relevant normal tissue (where toxicity occurs) ?
- Why do some patients respond better than others? How can we predict the best drug/combination for a particular patient ?
- How do cancers become drug resistant, and how can we combat this ?
- What is the effect of cancer drugs on immune cells and can drug combinations be used to enhance anti-cancer efficacy ?
- What are the most appropriate read-outs of drug efficacy and toxicity in clinical trials ?
This effort is multidisciplinary and inter-institutional involving investigators from the Department of Systems Biology at Harvard Medical School (Initiative in Systems Pharmacology), Harvard affiliated teaching hospitals and MIT.
- Miller MA, Chandra R, Cuccarese MF, Pfirschke C, Engblom C, Stapleton S, Adhikary U, Kohler RH, Mohan JF, Pittet MJ, Weissleder R. Radiation therapy primes tumors for nanotherapeutic delivery via macrophage-mediated vascular bursts. Sci Transl Med. 2017;9(392):eaal0225 – PMID: 28566423 – DOI: 10.1126/scitranslmed.aal0225
- Miller MA, Gadde S, Pfirschke C, Engblom C, Sprachman MM, Kohler RH, Yang KS, Laughney AM, Wojtkiewicz G, Kamal N, Bhonagiri S, Pittet MJ, Farokhzad OC, Weissleder R. Predicting therapeutic nano-medicine efficacy using a companion MR imaging nanoparticle. Science Transl Med. 2015;7:314 – PMID: 26582898
News
Please join us in congratulating Ralph Weissleder, MD, PhD and Matthias Nahrendorf, MD, PhD for being recognized as Highly Cited Researchers in 2025.
Clarivate Analytics’ Highly Cited Researchers List is an annual report of individuals at universities, research institutes and commercial organizations who demonstrate a significant and broad influence in their field or fields of research.
We're excited to share that the Mass General Brigham Research Spotlight features CSB (Weissleder Lab) paper published in Nature Biomedical Engineering, “Targeting immunosuppressive myeloid cells via implant-mediated slow release of small molecules to prevent glioblastoma recurrence."
Ralph Weissleder, MD, PhD is ranked #91 in the world and #62 in the United States among Best Scientists in Medicine for 2023.
The European Society for Molecular Imaging announced the winner of ESMI-2022: the society’s prestigious award is presented to Ralph Weissleder.

New PLoS Biology paper presents detailed, standardized citation data annotated for multiple impact indicators and their composite across science. Ralph Weissleder is in the top 0.01% of scientists based on their impact. Several other CSB PIs are in the top 1%. The raw data are available for download.