Miller Lab
Read BioLaboratory for Precision Therapeutics and Microenvironmental Dynamics
Our lab studies the functional dynamics of cells and their response to treatment from a quantitative, spatial, and multiscale perspective. We focus on inflammation and cancer, using a paradigm of image-guided pharmacology to characterize and engineer the behavior of therapeutics. With technologies like radiation-activated drug conjugates, ultrasound-responsive agents, and photoactivatable reporters, we spatially localize biological activity to targeted tissue while sparing off-target sites of potential toxicity.
Our work integrates quantitative imaging and computational modeling to dissect how multicellular regulation and stromal crosstalk unfold in tissue microenvironments and how they contribute to drug response or resistance. We quantify fluid dynamics and real-time therapeutic responses, matching drug delivery kinetics to spatial tissue context at a single-cell level.
Our work benefits from deep collaboration across the MGH Center for Systems Biology to marry computational analyses with advanced experimental models. We acknowledge funding from the NIH Common Fund, the National Cancer Institute, the Department of Defense, the American Thyroid Association, industrial partners, and others.
Join Our Team: We are actively recruiting in all project areas (Chemistry, Biology, Engineering) and review applications on a rolling basis. Please send CVs to miles.miller@mgh.harvard.edu.
Recent Publications
Di Ianni E, Jeon J, Mohammadi S, Hu H, Quintana JM, Lee C, Haidar EA, Goemans M, Zargani-Piccardi A, Mahamdeh M, Hernández IC, Rodriguez VE, Zhou Y, Aguirre A, Yuan S, Ng TSC, Breyne K, Lee H, Breakefield XO, Miller MA
Enhanced-mRNA Delivery Using Ultrasound-Delivered Anchors for Bioorthogonal Ligation. Angew Chem Int Ed Engl. 2026;:e23437 - PMID: 41568620 - DOI: 10.1002/anie.202523437Zhao P, Kundu J, Blanton D, Rezaeeyazdi M, Oudin MJ, Miller MA, Meyer AS, Bencherif SA, Baranov PY, Young MJ, Carrier RL
Human Retinal Progenitor Cell (hRPC) Migration in Three-Dimensional (3D) Environments of Varying Stiffness and Composition. J Tissue Eng Regen Med. 2025;2025:9963972 - PMID: 41195017 - PMCID: PMC12585844 - DOI: 10.1155/term/9963972Rodriguez-Castellanos VE, Quintana JM, Martin JM, Goemans MA, Banla LI, Ng TSC, Weissleder R, Miller MA
Radiation-Triggered Payload Release Enhances Bispecific Antibody Efficacy. Cancer Res. 2025;86(2):453-466 - PMID: 41066595 - DOI: 10.1158/0008-5472.CAN-25-0517Simpson GG, Quintana JM, Carrothers JE, Jiang F, Walker SA, Cho C, Weissleder R, Miller MA, Ng TSC
Fluorescent PSMA-Targeted Radiotheranostic Compounds for Multiscale Imaging. Bioconjug Chem. 2025;36(7):1448-1460 - PMID: 40574660 - PMCID: PMC12365983 - DOI: 10.1021/acs.bioconjchem.5c00139Obuchi W, Zargani-Piccardi A, Leandro K, Rufino-Ramos D, Di Lanni E, Frederick DM, Maalouf K, Nieland L, Xiao T, Repiton P, Vaine CA, Kleinstiver BP, Bragg DC, Lee H, Miller MA, Breakefield XO, Breyne K
Engineering of CD63 Enables Selective Extracellular Vesicle Cargo Loading and Enhanced Payload Delivery. J Extracell Vesicles. 2025;14(6):e70094 - PMID: 40527733 - PMCID: PMC12173531 - DOI: 10.1002/jev2.70094- More publications ...
Research projects
Engineering local drug delivery

Our research focuses on a key bottleneck in precision medicine: the disconnect between the molecular design of a drug and its actual behavior within a patient. Although next-generation bispecific antibodies, genetic therapies, and targeted small molecules have proven efficacy and hold transformative potential, their clinical utility is often capped by dose-limiting toxicities and insufficient uniform accumulation within diseased tissues. To address this issue, we engineer site-specific systems, ranging from radiation-activated conjugates to ultrasound-responsive agents, that respond to external triggers to release their payloads. Recent work includes radiation-activated antibody-drug conjugates and scintillating nanoparticle depots that convert ionizing radiation into local drug release. These promise to maximize local efficacy while minimizing systemic toxicity.
Radiation Therapy and the Tumor Microenvironment
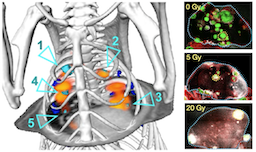
Radiation therapy is a cornerstone of cancer treatment, and the precision with which it can be delivered to tissues is continually improving with the development of conformal and adaptive treatments, molecularly-targeted radiopharmaceuticals, and new imaging capabilities. Our research investigates the impact of radiation on the tumor microenvironment, and in turn, how the microenvironment influences radiation response. A key objective is to use this understanding to identify and engineer new strategies for safely maximizing the effectiveness of radiation and radioconjugate treatments while using lower doses with improved safety.
Nanomaterials for localized therapy

Thousands of biomaterials and nanoparticles have been developed for drug and nucleic acid delivery, but molecular and cellular-level mechanisms of their action in tissue are often poorly understood. Our group expands on novel in vivo imaging techniques to analyze the combined distribution and action of biomaterials and nanoformulations at a multi-scale level. This project pursues a systematic, molecular-level understanding of drug action, with the goal of informing improved therapeutic and biomaterial designs.
Multi-scale dissection of tumor microenvironment dynamics
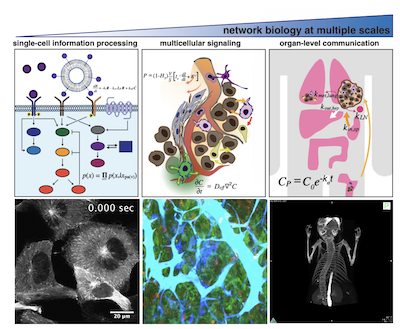
Through new technologies, including single-cell RNA sequencing (scRNAseq) and multiplexed tissue imaging (MTI), tissues can now be visualized with incredible molecular and cellular detail. However, such rich maps of tissue structure are typically static snapshots from a fixed sample, and lack important information about how the tissue actually functions — how cells, fluids, and biomolecules dynamically interact to govern multicellular behaviors. This project aims to overcome this limitation by building an integrated computational and experimental platform for quantitatively linking functional dynamics within tissue to a map of its molecular and cellular composition.