EXOSOME ANALYSIS PLATFORM
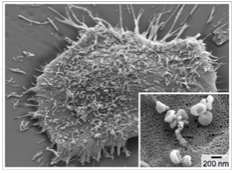
Exosomes. Scanning electron microscopy image of a primary human glioblastoma cell (GBM20/3), releasing abandant exosomes (inset).
Exosomes have emerged as a novel biomarker for clinical diagnostics. Exosomes are membrane-bound phospholipid nanovesicles (<200 nm in diameter) released into the circulation by cells. Exosomes present molecular markers, including those also expressed by their parental cells, and thus can serve as surrogates of parental cells. Quantifying exosomes from a specific cell type has been shown to have clinical importance; changes in subtypes of exosomes have been observed in patients with cardiovascular diseases, diabetes, infectious diseases, and cancer. Sensitive and selective exosome detection, however, still remains a challenging task, mainly due to the small size of these vesicles. Over the past years, my research has focused on developing new sensing technologies for quantitative exosome analyses. These efforts lead to new platforms that can 1) efficiently enrich exosomes from native specimen and 2) molecularly profile exosomes. Applying these platform, we unraveled a correlation between exosomal molecular profiles and disease progression, which validated the diagnostic potential of exosomes.
Microfluidics for exosome enrichment
Microfluidic filter for exosome separation. The device shown here uses sound waves to size-selectively enrich exosomes in flow condition.
We have developed on-chip filter systems to speed up vesicle separation from biological samples. For size-based separation, we have implemented an acoustic nanofilter that uses sound waves for force generation. Vesicles in an acoustic field experience a differential force proportional to their volume, and thereby can be fractionated according to their size. For molecular-specific enrichment, we adopted immunomagnetic selection to capture exosomes by their surface markers. A microfluidic device has been designed to perform magnetic enrichment as well as subsequent
RNA capture. Further device improvement is now under way to incorporate on-chip analysis capabilities.
nPLEX (nano-plasmonic exosome) sensor for high-throughput screening
nPlex system. An array of nanoholes are used as a substrate for surface-plasmon-resonance sensing. Exosomes bound to the sensor surface disturb plasmon waves. The first generation device (bottom right) has 36 detection sites.
We have developed a new surface-plasmon-resonance (
SPR) platform, termed nano-plasmonic exosome (nPLEX), that enables fast, high-throughput exosome protein profiling. The detection is based on extraordinary optical transmission through periodic nanoholes. The plasmonic nanohole array provides an ideal sensing scheme for exosomes: the probing depth (< 200 nm) of the sensor could be matched to exosome size to improve detection sensitivity; and the transmission optical setup can be used for detection, which allows for system miniaturization as well as the construction of highly-packed sensing arrays. We routinely operate nPLEX for high-throughput exosome screening. nPLEX chips with 100 detection sites are developed, and assay procedures have been automated.
iMEX (integrated magneto-electrochemical exosome) sensor
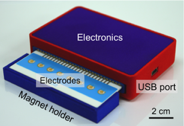
iMEX sensor. This prototype can simultaneously measure signals from eight electrodes. Small cylindrical magnets are located below the electrodes to concentrate immunomagnetically captured exosomes.
The iMEX (integrated magnetic-electrochemical exosome) was developed to allow for point-of-care exosome analysis. The iMEX integrates both vesicle isolation and detection into a single platform; target-specific exosomes are enriched through immunomagnetic selection, and subsequently detected by miniaturized electrochemical sensors. This strategy offers distinct advantages: 1) cell-specific exosomes can be isolated directly from samples without the need for extensive filtration or centrifugation; 2) the assay can achieve high detection sensitivity through magnetic enrichment and enzymatic amplification; 3) using the electrical detection scheme, sensors can be miniaturized and expanded for parallel measurements.