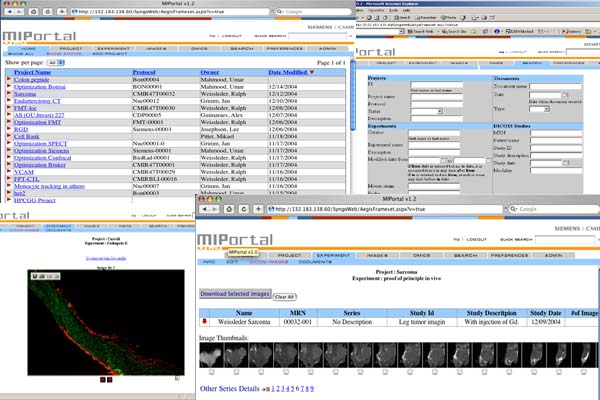
MIPortal
Small animal imaging systems can generate vast amount of data. For example, dedicated mouse MRI and CT systems generate an average of 1.5 GB of data per day, although peak data generation can be much higher (Paulus, Gleason et al. 2001). There is also a growing need to associate additional information with imaging studies, for example histology, immunohistochemistry, in situ hybridization, gels, microarrays or mass spectroscopy data. Furthermore, most research projects today require database queries and searches of protein structures (e.g. Entrez Protein), chemical data banks (e.g. Chembank, Pubchem) or other archives (e.g. Pubmed, CaBig). Finally, expanding multidisciplinary and multi-institutional research environments continue to foster collaborative projects, generating the need for organized and accessible shared data repositories. Typical information management systems, such as electronic laboratory notebooks or image archives, are either too cumbersome, limited to certain computer platforms, too slow or not fully adapted to handle the large number, types, and size of data files. Clinical data management systems based on PACS (Sinha, Bui et al. 2002) or electronic patient records (Sprague 2004) can be highly efficient but are prohibitively expensive for routine deployment in Molecular Imaging Centers and are not optimized to handle non-imaging associated data files. Given the above listed needs and shortcomings of commercial systems, we set out to develop a web-based information platform (MIPortal) with particular emphasis towards storage and retrieval of data sets. We initially defined our user criteria as follows:
- The system had to be web based and be compatible with common browsers operating on Macintosh, Unix or Windows clients
- The system had to be fast and hold large amount of data in real time with daily back-ups
- The system had to allow storage ofdifferent file formats (DICOM, TIff, .doc. xls. etc) with seamless integration, uploading, downloading
- The system had to allow for data analysis and storage of such analyzed data
- Be user friendly, intuitive to use, highly stable and follow the workflow of Molecular Imaging Centers.